Functools Partial
Partial application We have a function f(a,b) but we want to fix the values of b some value, and get a new function of a only. In pseudocode, we want to say g(a) = f(a,b=value). This is called a partial application of f.
Competition The most important competitor in python to functools.partial is the lambda function.
While there is other ways to create partial applications, I think these two methods are the most important ones.
In the below, I will compare functools.partial to lambda functions.
The differences
I will point out three advantages of functools.partial over lambda functions:
- Early binidng of arguments.
- Better metadata and
__repr__ - Compatible with
pickle
The final point is a more subjective one - I think functools.partial conveys intent more clearly than lambda functions do.
In small code bases, where the lambda is used as a literal and not a variable, this is less important, but as the code base grows, I think it is important to be able to read the code and understand what it does. So while I do use lambdas in my code, functools.partial is my preferred method of creating partial applications.
1. Early vs late binding of arguments
This code block is also a little longer, since I want to give a full example of how to use functools.partial and lambda functions.
import scipy.integrate
opts = dict(mass=1, spring_constant=1, viscous_damping_coefficient=1, t_span=[0, 10], y0=[1, 0])
def hook(t, y, k, m, c):
"""Damped mass-spring oscilllator model
m * acc = - k * pos - c * vel
"""
pos,vel = y
return [vel, - k * pos / m - c * vel / m]
# solve_ivp requires a function of the form f(t,y), so we can use a lambda to pass in the side parameters
hook_lambda = lambda y, t: hook(y,t,k=opts['spring_constant'],m=opts['mass'], c=opts['viscous_damping_coefficient'])
out1 = scipy.integrate.solve_ivp(hook_lambda, t_span=opts['t_span'], y0=opts['y0'])
# but it uses late binding! so this can be surprising
# the lambda will pick the "most recent" value of opts
opts['mass'] = 2
out2 = scipy.integrate.solve_ivp(hook_lambda, t_span=opts['t_span'], y0=opts['y0'])
# one way to avoid this is to use a functools.partial
# it uses early binding, so the values are "baked in" when the partial is created, not when it is called
import functools
opts['mass'] = 1
hook_partial = functools.partial(hook,
k=opts['spring_constant'],
m=opts['mass'],
c=opts['viscous_damping_coefficient'])
out3 = scipy.integrate.solve_ivp(hook_partial, t_span=opts['t_span'], y0=opts['y0'])
# again, change the mass and run again
opts['mass'] = 2
out4 = scipy.integrate.solve_ivp(hook_partial, t_span=opts['t_span'], y0=opts['y0'])
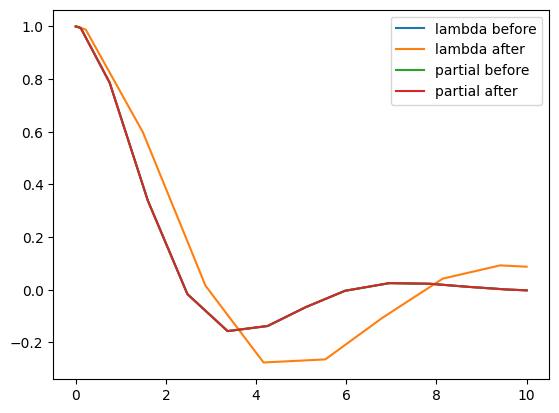
# plot the four solutions, for comparison
# only the line 'lambda after' differ from the others
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.plot(out1.t, out1.y[0], label='lambda before')
ax.plot(out2.t, out2.y[0], label='lambda after')
ax.plot(out3.t, out3.y[0], label='partial before')
ax.plot(out4.t, out4.y[0], label='partial after')
plt.legend();
2. The metadata differ
# They have different print outputs
print(hook_lambda)
print(hook_partial)
# and there is a lot of useful metadata on the partial that the lambda doesn't have
print(hook_partial.keywords)
print(hook_partial.func)
<function <lambda> at 0x7ff176eee5f0>
functools.partial(<function hook at 0x7ff176eee680>, k=1, m=1, c=1)
{'k': 1, 'm': 1, 'c': 1}
<function hook at 0x7ff176eee680>
3. Partials work with the pickle module, but lambdas don’t.
This shows up in machine learning sometimes e.g. when serializing some model or metadata from a training process. It is also the prerequisite for using partials with multiprocessing.
import pickle
pickle.dumps(hook_partial)
print("Partial pickles successfully")
try:
pickle.dumps(hook_lambda)
except pickle.PicklingError:
print("Failed to pickle the lambda")
Partial pickles successfully
Failed to pickle the lambda
